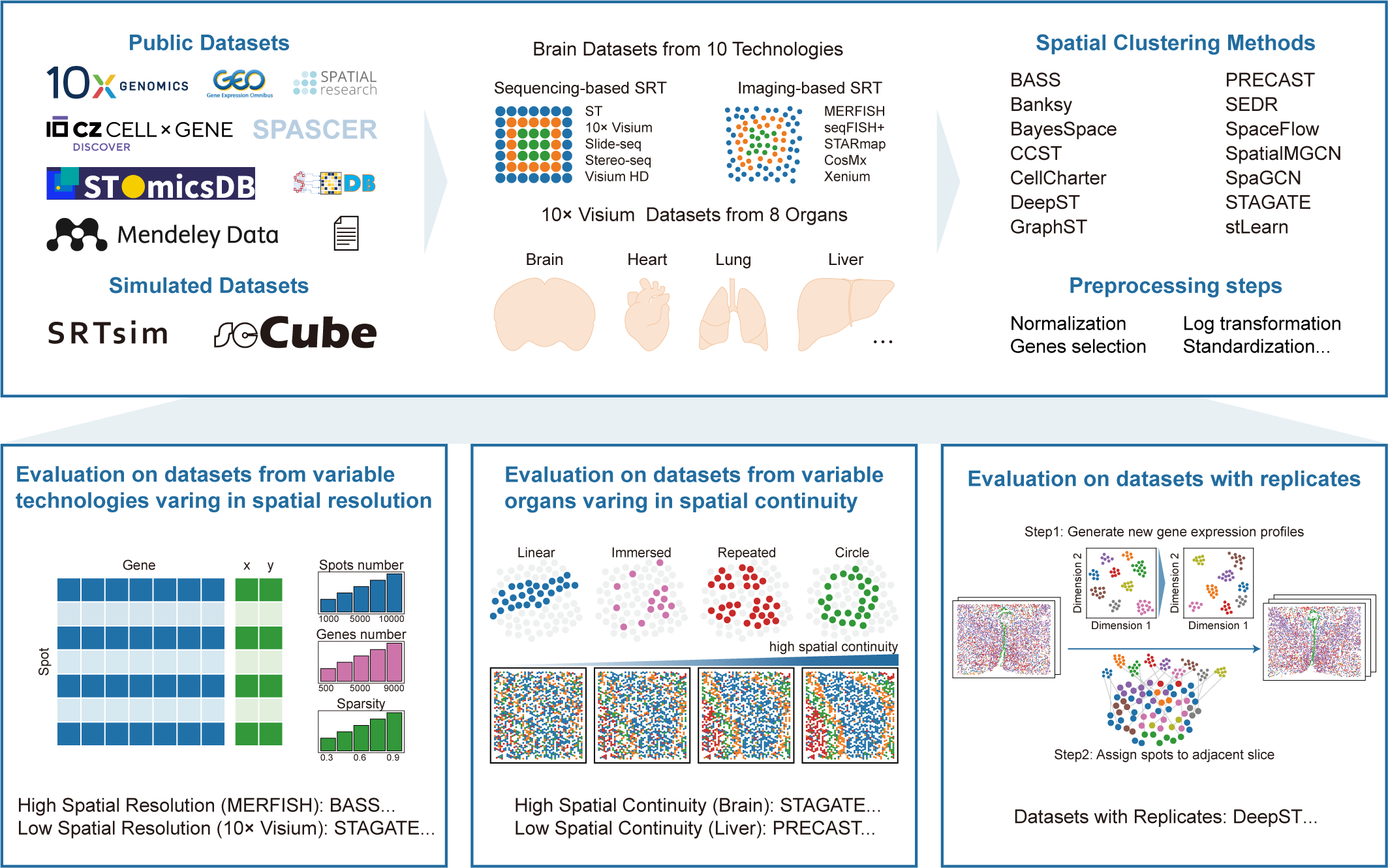
Spatial clustering is a critical step in the analysis of spatially resolved transcriptomics, serving as the foundation for downstream investigation of tissue heterogeneity. Although numerous computational tools have been developed, systematic benchmarking across different technologies, organs, and biological replicates has been limited. Here, we present a comprehensive evaluation of 14 spatial clustering methods using approximately 600 datasets, including both real-world and simulated data with ground truth. We evaluated accuracy and applicability across diverse technologies and organs, revealing method-specific strengths and preferences. Using simulation of adjacent tissue slices and spatial neighborhood disruptions, we further examined performance in the context of biological replicates. Furthermore, we investigated how data characteristics, spatial distribution patterns, and preprocessing pipelines influence clustering outcomes. Together, our results provide practical benchmarking guidance, enabling researchers to select appropriate spatial clustering methods tailored to specific technologies, organs, and biological replicates.